Exploring the microbiome's impact on immunotherapy efficacy

In our previous blog post (check it out here), we explored how the microbiota can have a profound impact on our body’s systemic immune response. Did you know that the diversity and identity of your gut microbiome can affect responsiveness to immunotherapy?
This raises an interesting question: can we use the types and numbers of the species living in your gut and the metabolites they produce to predict how well you will respond to certain therapies? In the context of cancer immunotherapy, studies have shown that the presence or absence of specific microbes can be associated with better anti-tumor T-cell responses following cancer therapy. Indeed, in a mouse model, anti-CTLA4 antibody treatment was effective in controlling tumor progression only in specific pathogen-free animals, not in germ-free animals (animals raised without microbes). But then, when they introduced distinct Bacteroides species in these germ-free animals, the expected effects to the antibody treatment were restored. Anti-tumor effects were perceived by the introduction of species Bifidobacterium along with anti-PD1 antibody treatment against melanoma. Amazing right?
Well, currently the field of the microbiome at the intersection of immunotherapy is a very active field of research and some microbiome-targeted therapeutic strategies have been successfully implemented. For example, fecal microbiota transplantation from response donors has shown beneficial changes in immune cell infiltrates and gene expression profiles in both the tumor microenvironment and the gut lamina propria.
The microbes matter, as well as their metabolites that have significant impacts on immunotherapy responses. For instance, short-chain fatty acids (SCFAs) have been shown to have suppressive effects on tumor cells, with higher SCFA concentrations associated with longer-term therapy responses and longer progression-free survival. Also other metabolites can affect immunotherapy responses either shaping a tumor-suppressing microenvironment, such as secondary bile acids, inosine, kynurenine, butyric acid or a tumor-promoting microenvironment such as polyamides, lipotheichoic acid and ketone.
Ongoing studies are using metagenomics sequencing techniques, along with metabolomics techniques, to evaluate the potential of the microbiome as a biomarker of patients' responsiveness to immunotherapy. The gut microbiome and metabolome are becoming increasingly critical research targets in the context of cancer immunotherapy. Exciting times lie ahead!
How can intestinal microbiota and its metabolites influence immunotherapy?
So, I think we agree that understanding how particular gut bacteria can affect the immune system is crucial. The microbes can determine the outcome of immune therapy, specifically immune checkpoint inhibitors. They do that by affecting the immune system in two ways: antigen-independent and antigen-specific.
For the first mechanism, certain bacteria can indeed influence the differentiation of CD4 T-cells into subsets of different immune cells impacting the final immune response. But these bacteria can also send signals and activate innate immunity via dendritic cells or macrophages providing an extra level of immune support. There is this probiotic, Lactobacillus rhamnosus, that was shown to promote innate immune responses in combination with anti-PD1 antibody therapy by activating dendritic cells. This highlights the potential for microbiome-based therapeutics such as probiotics (“live”. bacteria), prebiotics (source of food for your gut healthy bacteria), but also fecal microbiota transplantation and neutraceuticals to support our immune system in various ways.
Second, in the antigen-specific mechanism, you have certain bacterial species that can trigger cross-reactive anti-tumor responses. By expressing certain antigens, they “mimic” the tumor neoantigens (antigen mimicry) that are targeted by T-cells and thus can stimulate T-cell amplification enhancing the immune response to cancer cells. Which is exactly what we want! Cool, no? This is relevant in immunotherapy, where targeting neo-antigens that arise from mutations in tumor cells is critical for the success of treatment. By the presence of these microbial antigens, a cross-reactive anti-tumor response is triggered. This microbial antigen-cancer cross-reactivity is very relevant for identifying therapeutic strategies and improving patient outcomes.
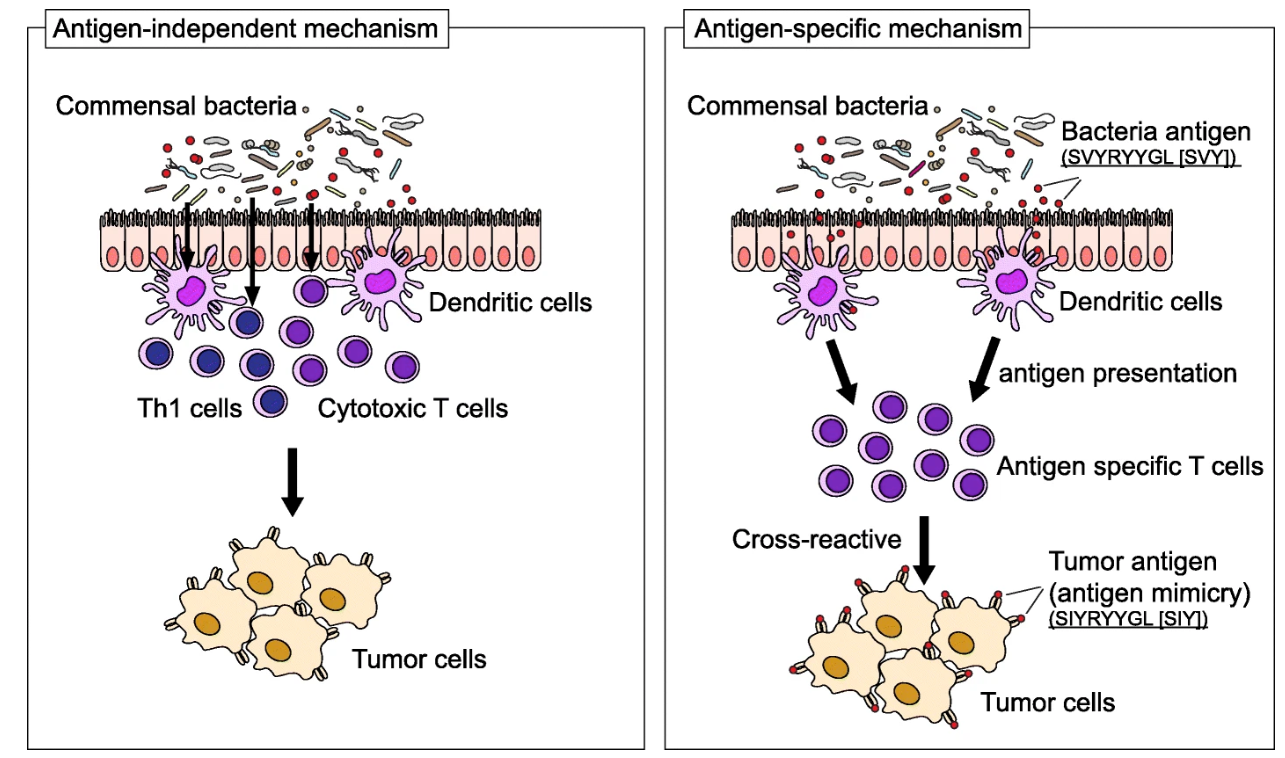
Source | Increased microbial diversity and the presence of specific species are associated with your immune response against the tumor, either beneficial or detrimental. Antigen-dependent mechanisms (left): specific immune responses are induced by commensal bacteria; Antigen-specific mechanisms (right): these appear by the presence of a specific epitope expressed in a particular species, for e.g. Bifidobacterium breve, T-cells react to these epitopes and cross-react with a neo-antigen (similar antigen). Cross-reaction between commensal bacterial and tumor antigens appear to be relevant in immune checkpoint inhibitor therapy.
In the context of antibody-based therapeutics, two questions appear to be relevant: (1) Can the gut microbiome profile be used as a biomarker to differentiate responders from non-responders in antibody therapy? (2) How can we put the microbiome in the context of antibodies?
The gut microbiome as a predictive biomarker for immunotherapy responsiveness
Machine learning (ML) methods are popular, and yes also for microbiome-based predictions! Microbiome data have been used for predicting responses to immunotherapy. In a recent meta-analysis study, 16S rRNA taxonomic variables and community diversity indexes were used to train and validate machine-learning models. Hierarchical clustering analysis revealed a higher response rate among patients with increased abundances of the Phylum Firmicutes, while lower response rates for patients with increased abundances of the Phylum Bacteroidetes. The ML models developed by the researchers to predict immunotherapy response status using microbiome features showed an area under the curve (AUC) of 0.75, indicating good predictive performance and more interestingly, they showed cross-sequencing platform applicability suggesting the results are generalisable to broader datasets. Nice study!
Of course, there is still plenty more research going on regarding ML models to unlock the microbiota’s potential to boost immunotherapy. This is not without any challenges. Microbiome datasets typically have high dimensionality with relatively low sample sizes which leads to the statistical problem called the curse of dimensionality. Indeed, we have typically a very large number of taxonomic features while only a small number of feces samples. Luckily there are some tricks to deal with this cacophony of data,. For example, we can calculate the so-called alpha-diversity which summarizes the number of taxonomic features in a given sample (i.e. a person’s gut microbiome). The beta-diversity, on the other hand, describes how the diversity of gut microbiomes can be compared between patients. A recent deep representation learning framework called DeepMicro was capable of compressing microbial features that could successfully predict various disease outcome.
To accurately predict immunotherapy response, a patient's microbiota profile ideally should be combined with other predictive markers such as tumor mutation load, geographical factors, tumor type, and dietary variables. Therefore, researchers aim to integrate multi-omics levels in a multilevel and multidimensional research design towards personalized antibody therapy in the future. Despite challenges, the potential benefits of using machine learning to predict immunotherapy response based on gut microbiome data make it an exciting area of ongoing research.
Restoration of the gut microbiome: the intersection of antibodies and the microbiome
Maybe you are already convinced that the microbiome might be a game-changing breakthrough in revolutionizing immunotherapy treatments? For those who aren't … let’s dive in a bit deeper!
Did you know that there are clinical trials underway to evaluate the combination of microbiome therapeutics, such as probiotics and FMT, and immunotherapy, opening up exciting new possibilities for cancer treatment? One such trial is exploring the use of Bifidobacterium species in combination with pembrolizumab, an anti-PD1 antibody, for patients with advanced metastatic colorectal carcinoma, triple-negative breast cancer, and checkpoint inhibitor relapsed tumors. The combination therapy has the goal to bring beneficial immunomodulatory activity. The implications of this microbiome-immune research can be enormous, and the possibilities are endless. By manipulating the microbiome, we may be able to unlock the secrets of the immune system and revolutionize cancer treatment. So stay tuned, because the future of cancer treatment is now, and the microbiome might take the lead.
… But what can the microbiome bring to improve antibody therapy?
Well, first we take into account microbiome features because bacteria might affect the reactivity of T-cells. Recent research has shed light on this molecular mimicry that we touched upon in the beginning of this blog. It occurs between the microbiota and the antigens that T-cells react to. A study analyzing epitopes from 38 antigenic categories found that the sequence similarity of various antigens to those presented by the microbiota can either decrease or increase T-cell reactivity. The genus from which the microbiota homologous sequences were derived could even determine whether a tolerogenic or inflammatory effect was observed. This research suggests that the presence or absence of microbiome sequences can contribute to the toleration or reactivity of T-cells. Yet another study showed that some bacterial peptides could modulate the immune response of human peripheral blood mononuclear cells in vitro, related to higher Th17 and Th22 responses. Yes, indeed, bacterial peptides seem to be inductors of human immune responses! wow!
The repertoire of bacterial species in the gut might show potential similarities with tumor antigens. And they might be presented to immune cells as well! This goes together with co-signals that allow functional responses or maturation of subset of cells. So, it makes sense to identify bacterial components that share similarities with human tumor antigens because your immune system has already “seen” these antigens earlier (I hope you do remember this from the first blog on the microbiome-immune axis?). You might have built an immune response against the microbiota sequence variant where immune cells will be reactivated by encountering the human tumor-related antigenic epitope. Thus, this re-encountering strengthens the anti-tumoral response and increases therapeutic efficacy. For the first time, a recent study showed high homology between peptides derived from microbiota species of the Firmicutes and Bacteroidetes phyla and tumor-associated antigens. The study suggests the anti-microbiota T-cell memory might turn out an anti-cancer T-cell memory that controls cancer growth when the expressed tumor-associated antigen is similar to the microbiota epitope. Indeed, it looks like that by keeping your microbiome as diverse as possible, cancer patients might have a selective advantage.
Conclusion
You may understand now that the microbiome is a complex ecosystem that plays a crucial role in maintaining our health and well-being. The microbiome seems to have beneficial effects on our immune system and in case of illness, it even can improve disease outcome. Incorporating microbiome features into immunotherapy is thus a promising approach that may enhance its effectiveness. Various clinical trials are underway that evaluate the combination of microbiome-therapeutics and immunotherapy such as fecal microbiota transplantation, live biotherapeutic products, or pro/prebiotics supplementation.
As we continue to unravel the mysteries of the immune system along, we may discover even more innovative approaches to treating diseases and improving overall immune health. But one thing is clear: protecting and nurturing our microbial friends could be the key to unlocking a new era of cancer treatment.
Sources:
- https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-021-00923-w
- https://pubmed.ncbi.nlm.nih.gov/26541610/
- https://pubmed.ncbi.nlm.nih.gov/26541606/
- https://clinicaltrials.gov/ct2/show/NCT03643289
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9797511/pdf/fphar-13-1091124.pdf
- https://gut.bmj.com/content/71/3/521
- https://www.oncotarget.com/article/28252/text/
- https://reader.elsevier.com/reader/sd/pii/S1438422122000133?token=94E94DA9C07921D888C967849D2A7420236B883714ECF5D69B37DBBF99529ACB6D8A6895FF2DBFB91786F1F7DB9DF49F&originRegion=eu-west-1&originCreation=20230220092359
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6604038/
- https://clinicaltrials.gov/ct2/show/record/NCT03775850
- https://www.esmoiotech.org/article/S2590-0188(20)30013-7/fulltext
- https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0196551
- https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-022-03512-6
- https://www.frontiersin.org/articles/10.3389/fmicb.2017.01726/full
- https://www.cimd.fraunhofer.de/en/projects/antibody-mediated-microbiome-correction.html
- https://www.science.org/doi/10.1126/sciimmunol.abg3208
- https://www.nature.com/articles/s41598-020-63159-5
Subscribe to our blog:






